Research
At ChlamHealth, we study how microbes interact, adapt and spread — from sexually transmitted infections to zoonotic diseases. Our goal: to create knowledge and tools that improve prevention, diagnosis and responsible treatment in a One Health perspective.
Interactive 3D Chlamydia
Explore this interactive 3D model by moving your mouse to gently orbit the scene.
Click on the floating labels (e.g. human, fish, koala) to open related research papers in a new tab.
Note for screen reader users: This content cannot be displayed in an accessible format.
All related research papers are provided as text links below.
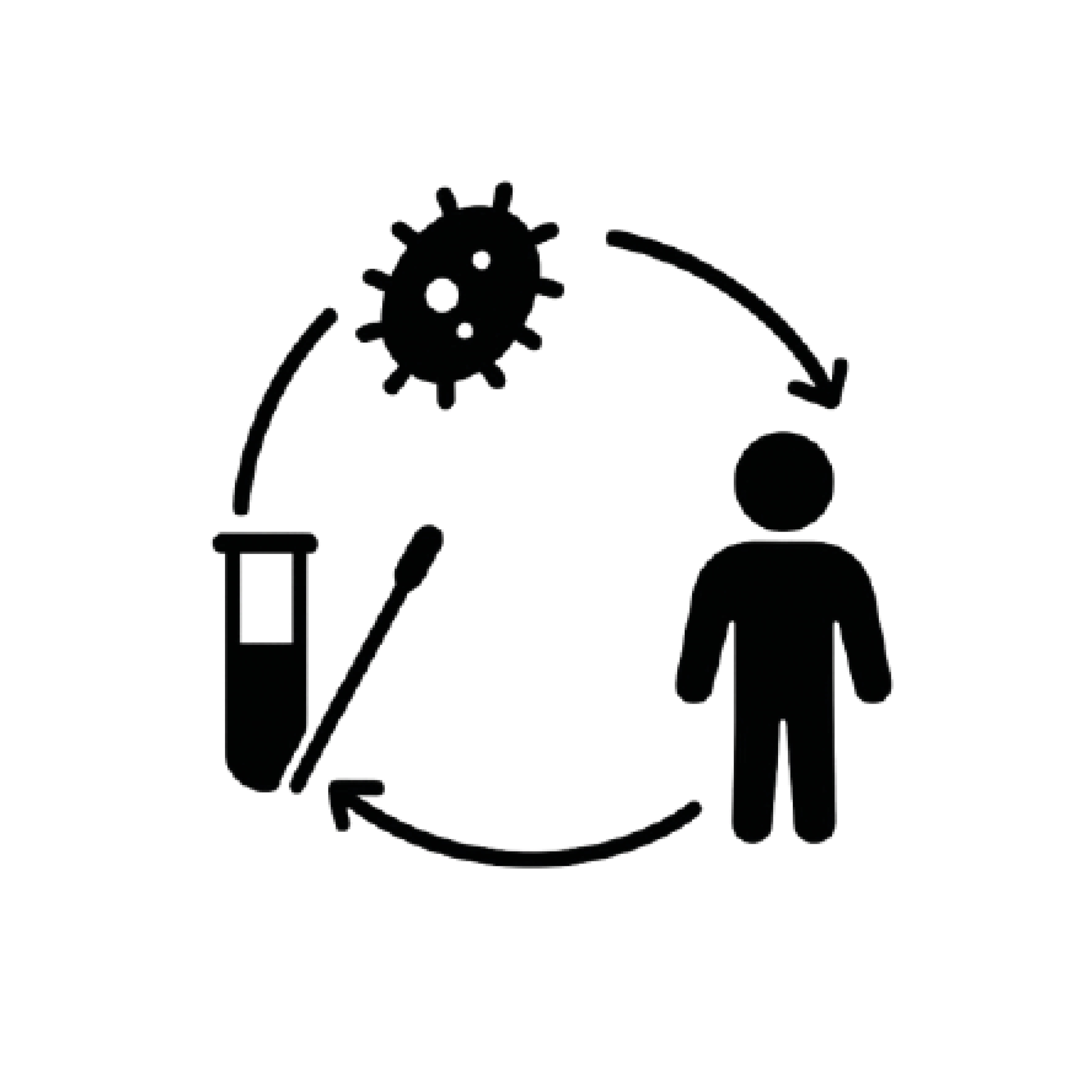
The STI Interactome
Nicole Borel, Enrique Rayo, Delia Onorini
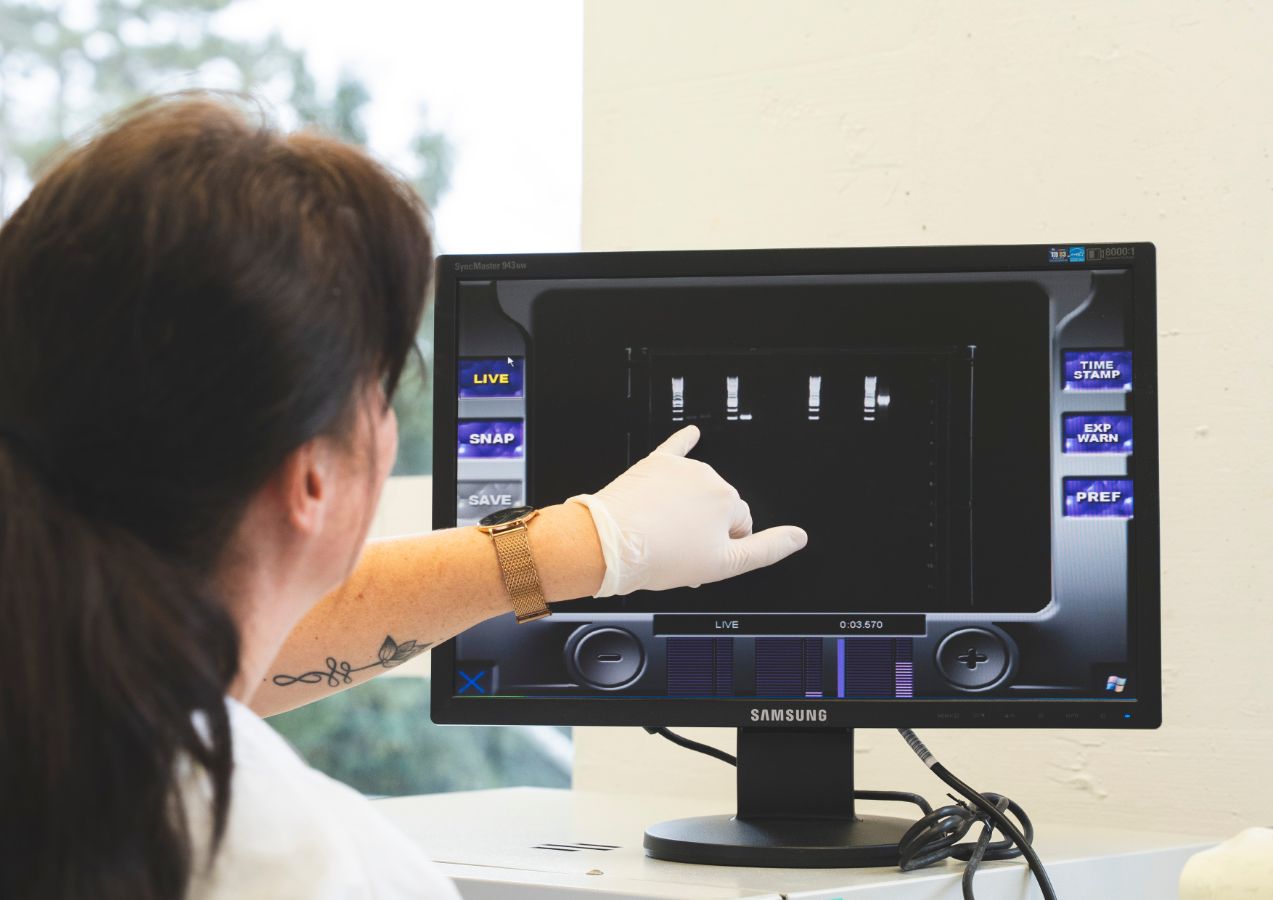
The STI interactome Project investigates the complex network of interactions between sexually transmitted pathogens, commensal microbiota, and antimicrobial resistance mechanisms within the urogenital and rectal environments. By combining culturomics, metagenomics, and pathogen genomics, the project aims to map the ecological and functional relationships that define the STI interactome. Special attention is given to co-infections, microbial competition, and the selective pressures introduced by antimicrobial treatments, with the goal of advancing our understanding of STI persistence, transmission, and treatment response in diverse populations.
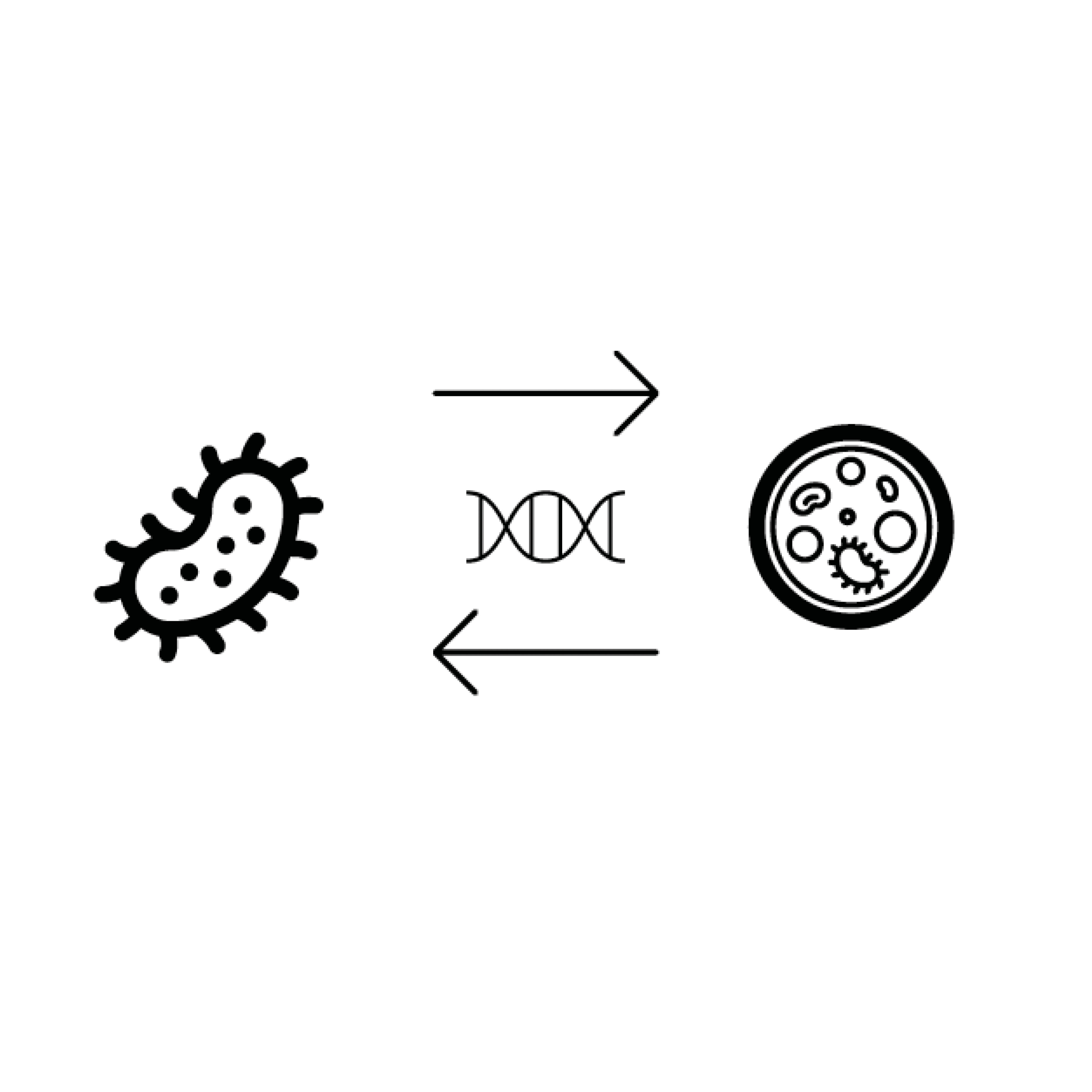
The Microbial Mobilome
 Hanna Marti
Hanna Marti
The Microbial Mobilome Project investigates the frequent exchange of antimicrobial resistance determinants and virulence markers among bacteria. This exchange is facilitated, in part, by rapid evolutionary mechanisms that include transduction, conjugation and transformation, driven by bacteriophage infection, plasmid-donor-recipient interaction and bacteria-owned competence machineries, respectively. We aim to unravel this complex microbial mobilome with a particular focus on facultative and obligate intracellular Gram-negative bacteria and their specific DNA exchange mechanisms.
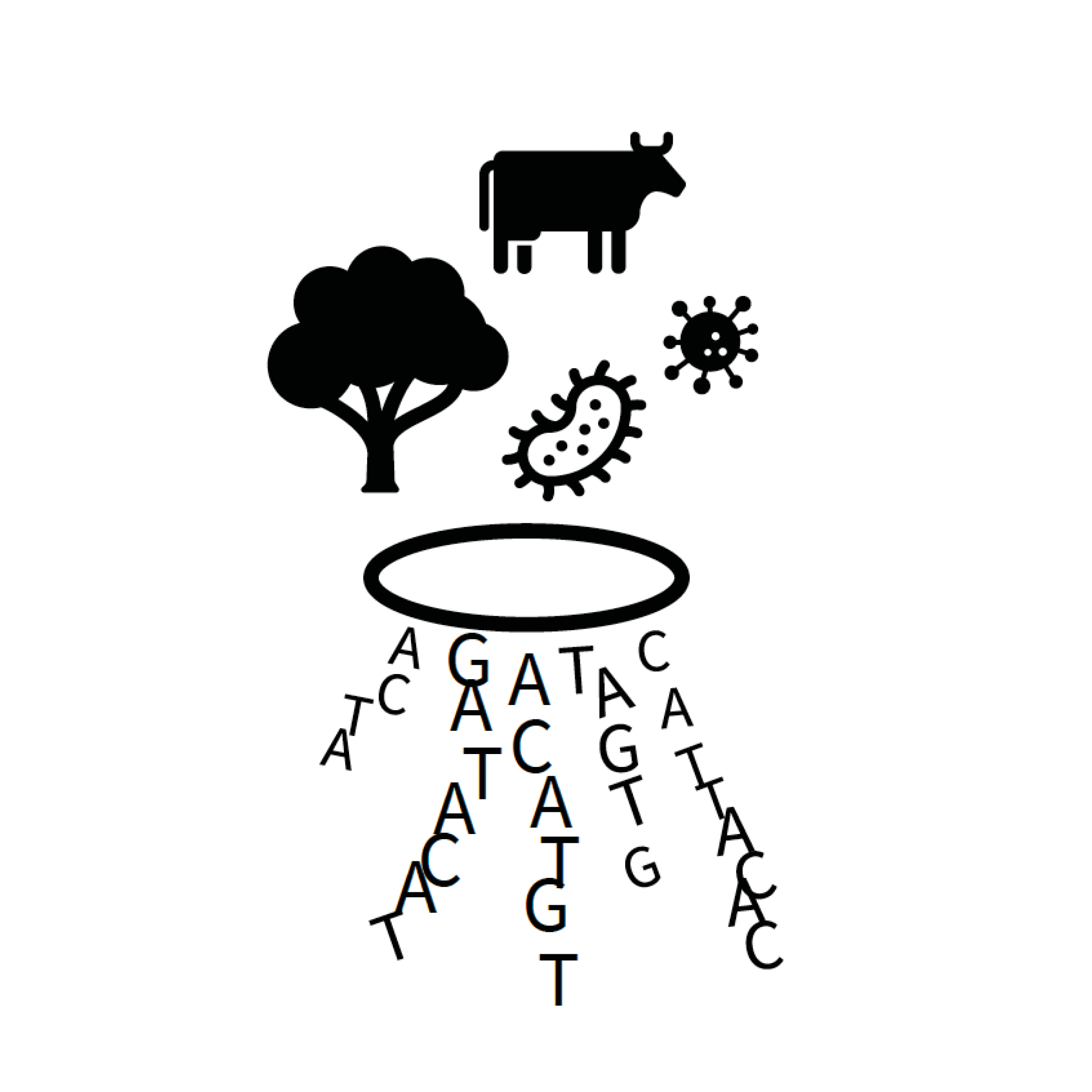
The Environmental Pathobiology
 Nicole Borel, Hanna Marti, Enrique Rayo
Nicole Borel, Hanna Marti, Enrique Rayo
The Environmental Pathobiology Project takes a step beyond host-host, host-pathogen and pathogen-pathogen interaction by investigating the environment as a factor for microbial fitness and viability. This project takes a holistic approach to elucidate the influence of the environment on the interactome between hosts, pathogens and microbial communities. Our specific aims are to identify environmental factors that influence bacterial viability and to develop molecular tools for rapid distinction between live and dead bacteria in complex samples.